 Last Thursday, a research group in Singapore has successfully obtained US$5 million (S$7.7 million) from the US National Institutes of Health (NIH) to sequence the DNA of the Elephant Shark.
Last Thursday, a research group in Singapore has successfully obtained US$5 million (S$7.7 million) from the US National Institutes of Health (NIH) to sequence the DNA of the Elephant Shark.This is a collaborative effort between the Institute of Cell and Molecular Biology (IMCB) in Singapore and the J. Craig Venter Institute in the USA.
Associate Professor Byrappa Venkatesh is the principal investigator of the Singapore team. They spent more than two years searching the world for a suitable shark species.
The project is already well underway and Prof. Venkatesh estimates that it will take up to two years to complete.
You might be thinking: "Almost 8 million dollars for a shark genome? WHY???"
Is it a particularly pretty shark?

Heh, not really. With funny looking teeth, buggy eyes and an ugly snout, the elephant shark is not much in the looks department.
Just compare it to the graceful curves of an elegant blue shark below.

What a striking contrast!
The Straits Times reported that the purpose of the project is to learn more about gene function and improve our understanding of human diseases.
That is the long-term goal of many genome projects but not the main reason why this bizarre-looking shark species was chosen. There were no details about the evolutionary significance of this project.
It was also inaccurately reported that sharks were the first creatures to develop nervous and immune systems (invertebrates eg. insects already have those, just that sharks have immune systems that are relatively closer to humans).
Not many fun details from a newpaper report!
So, based on a lecture given by Prof. Venkatesh in January 2007, Fresh Brainz is delighted to answer the biggest question on your mind:
What's so frickin' important about this ugly shark?
1. Outgroup Species To Study Fast Teleost Evolution
This may sound strange, but fish are the largest group of vertebrates. There are over 28 000 species of fish, of which about 27 500 are ray-finned fish. In comparison, all the other vertebrates (including human beings) combined add up to only about 24 000 species.

Ray-finned fish are interesting to scientists because they have evolved a wide variety of shapes and sizes. This is especially true for an infraclass of ray-finned fish called teleost fish. This rapid diversification of body forms is called the teleost radiation.
How did the teleosts evolve so quickly?
One possibility is because of a whole genome duplication event that happened approximately 350 million years ago. Teleost fish have duplicate copies of many genes compared to other fishes, and even mammals such as us.
For example, mammals have 4 sets of Hox genes, whereas teleost fish have 7 sets (2X4 sets with one set loss). This increase in genomic information may have provided the raw materials for fast teleost evolution.
Scientists already have genomic information from teleost fishes such as the zebrafish and fugu, but in order to discover teleost-specific features they need an outgroup species for comparison.
Cartilaginous fish, such as sharks, are good candidates as outgroup species because they occupy an appropriate position on the evolutionary tree.
Also, sharks don't appear to have undergone a genome duplication event. In fact, sharks evolve so slowly that their DNA substitution rate is 7 times slower than other fish. This may be one main reason why sharks appear relatively unchanged for hundreds of millions of years.
2. Smallest Shark Genome
OK, so it's important to go get some shark sequence. But which shark should it be?
Unfortunately, preliminary data showed that the genome sizes of well-known sharks tend to be much larger than the genomes of human beings. This means that the sequencing effort would involve a lot of time and money.
So Prof. Venkatesh and his colleagues searched the globe for any rare shark species that has a more compact genome. Eventually they tested the elephant shark (Callorhinchus milii) which is native to the waters of south Australia and New Zealand.
Early estimates of its genome size was about 1.2 billion base pairs of DNA (compared to human 3.2 billion bp). Later as the sequencing work progressed this figure was revised to only 910 million bp, the smallest genome of any known shark.
This made the elephant shark the most attractive shark model available.
3. Surprisingly Similar Genomic Structure To Humans
Although sharks are more distantly related to humans than teleosts, sequencing results revealed that they have a surprisingly closer genome structure to us.
This was discovered by comparing regions of synteny between sharks and humans. If a particular set of genes had exactly the same arrangement in both sharks and humans, then that region is called a syntenic region.
Current data indicates that syntenic regions are more conserved between humans-and-sharks, compared to between humans-and-teleosts.
Thus, the elephant shark genome project is a useful resource for the detailed investigation of human genomics. It can also be used to study the genomes of many other vertebrate animals.
Not too shabby for such an ugly shark.
Not too shabby.
Would you like to know more?
- Elephant shark genome project website
- Choosing an appropriate model organism





 Science policy analyst Cong Cao, who worked in Singapore before, wrote a letter to Science magazine (8 June 2007), listing a number of challenges that the Singaporean scientific endeavour faces.
Science policy analyst Cong Cao, who worked in Singapore before, wrote a letter to Science magazine (8 June 2007), listing a number of challenges that the Singaporean scientific endeavour faces. Aww! What a cute puppy!
Aww! What a cute puppy!
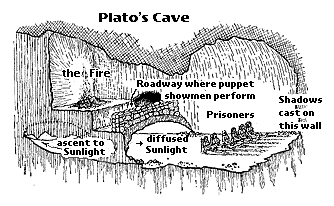
 After Plato, another philosopher's idea was to become a major setback to the development of modern scientific thinking.
After Plato, another philosopher's idea was to become a major setback to the development of modern scientific thinking.







 Fresh Brainz is proud to bring you yet another collection of my favourite original photos.
Fresh Brainz is proud to bring you yet another collection of my favourite original photos.













 Watch
Watch 
 I accidentally bit myself last week while stuffing my face with a bowl of fishball noodles.
I accidentally bit myself last week while stuffing my face with a bowl of fishball noodles.







